Reference
Ryan BC, Werner TS, Howard PL, Chow RL. (2016) ImiRP: a computational approach to microRNA target site mutation. BMC Bioinformatics 17:190.
Welcome to the Illegitimate microRNA Predictor (ImiRP), an open source Web application designed to selectively mutate predicted miRNA target sites while ensuring that predicted target sites for other miRNAs, termed illegitimate target sites, are not created in the process. Accidental creation of illegitimate target sites upon disruption of existing miRNA target sites can complicate analysis of miRNA-target regulation. Given the small size of miRNA recognition motifs and the abundance of identified miRNAs, the likelihood of generating an illegitimate site upon mutation of an existing site is high. This becomes even more problematic when trying to mutate multiple predicted target sites simultaneously.
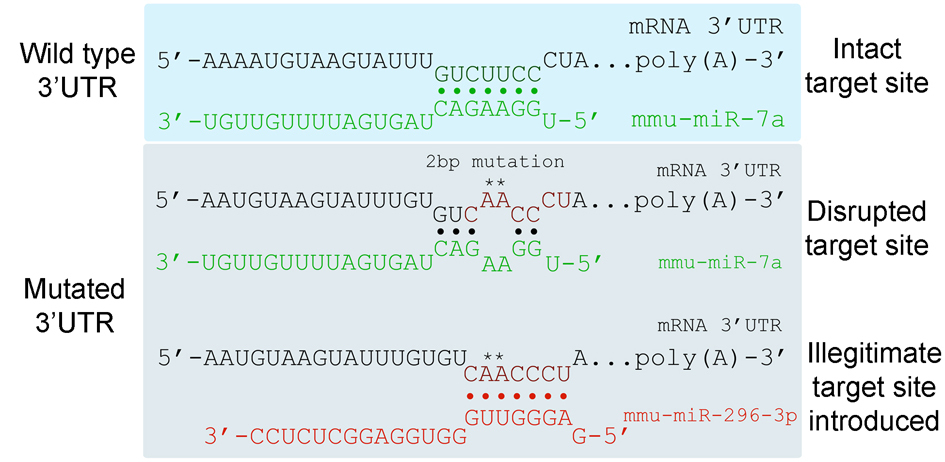
ImiRP is a tool that enables researches to input their favourite 3’UTR DNA or mRNA sequence and specify the species of interest and locations of miRNA target sites they wish to mutate. ImiRP automates the process of specified target site mutagenesis, and identifies and displays up to five mutations that do not generate illegitimate sites. This tool is especially powerful when investigating cooperative regulation of a single transcript by multiple miRNAs, where multi-site mutagenesis may be necessary.
All mature miRNA sequence information was provided by the miRBase version 21 high confidence miRNA dataset.
Ryan BC, Werner TS, Howard PL, Chow RL. (2016) ImiRP: a computational approach to microRNA target site mutation. BMC Bioinformatics 17:190.